Dated mammal phylogeny from Phillips et al. (PNAS, 2009), and carpet python (Morelia spilota) on field trip to Cunningham's Gap
Research areas
Mammalian Evolution

1. What on Earth are monotremes? – tracing their evolution from “normal” ancestors.
2. Reconstructing marsupial evolution.
3. Dating the major diversification of placental mammals – testing links to the dinsosaur extinction and the "Cretaceous terrestrial revolution" .
4. Are marsupials intrinsically inferior to placentals (as Lillegraven suggested) or does it come down to evolutionary context differences?
Avian Evolution
Phylogenetics and molecular dating
1. Modelling morphological evolution for merging fossils into the tree of life.
2. Total evidence dating (combined molecular-morphological matrices) is interesting, but when is it useful?
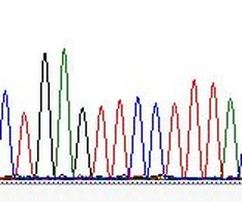
3. Identifying and ameliorating DNA substitution model misspecification for inferring phylogeny and molecular dates.
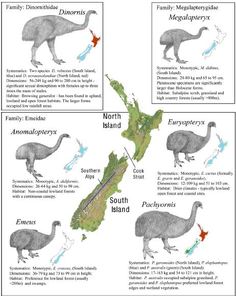
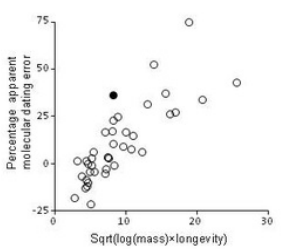
Palaeobiology
Some collaborators on recent grants /papers
Andrew Baker (QUT)
Robin Beck (University of Salford)
Simone Blomberg (University of Queensland)
Carmelo Fruciano (CNR, Italy)
Susan Fuller (QUT)
Gillian Gibb (Massey University, NZ)
Daniel Ksepka (Bruce Museum)
Simon Ho (University of Sydney)
Michael Lee (University of Adelaide)
Maria Nilsson-Janke (Senckenberg Museum)
Paul Oliver (Queensland Museum & Griffith University)
Peter Prentis (QUT)
Gavin Prideaux (Flinders University)
Kenny Travouillon (Western Australian Museum)
Vera Weisbecker (Flinders University)
Robin Beck (University of Salford)
Simone Blomberg (University of Queensland)
Carmelo Fruciano (CNR, Italy)
Susan Fuller (QUT)
Gillian Gibb (Massey University, NZ)
Daniel Ksepka (Bruce Museum)
Simon Ho (University of Sydney)
Michael Lee (University of Adelaide)
Maria Nilsson-Janke (Senckenberg Museum)
Paul Oliver (Queensland Museum & Griffith University)
Peter Prentis (QUT)
Gavin Prideaux (Flinders University)
Kenny Travouillon (Western Australian Museum)
Vera Weisbecker (Flinders University)
Scripts/programs
Genbank_pl is a perl script for extracting rRNA, tRNA and protein coding DNA sequences from mitochondrial genome accessions (Scott Roy, Tim White, Matt Phillips).
NicheSim is a C++ program that simulates the evolution of discrete characters in 2D niche (or morphological) space on a phylogeny. It is non-Markovian, in that evolutionary transitions can be influenced by state history or by states in other contemporaneous taxa. Several simple metrics of ecomorphospace occupation are also calculated for the simulated or observed data. (Tim White, Matt Phillips).
Genbank_pl and NicheSim are available from Matt Phillips upon request.
NicheSim is a C++ program that simulates the evolution of discrete characters in 2D niche (or morphological) space on a phylogeny. It is non-Markovian, in that evolutionary transitions can be influenced by state history or by states in other contemporaneous taxa. Several simple metrics of ecomorphospace occupation are also calculated for the simulated or observed data. (Tim White, Matt Phillips).
Genbank_pl and NicheSim are available from Matt Phillips upon request.